Size of protein aggregates, not abundance, drives spread of prion-based disease
Size of protein aggregates, not abundance, drives spread of prion-based disease
In a study that challenges the conventional wisdom about infections caused by proteins called prions, Brown researchers report in Science that the size of protein structures, rather than their abundance, determines their transmission among cells.
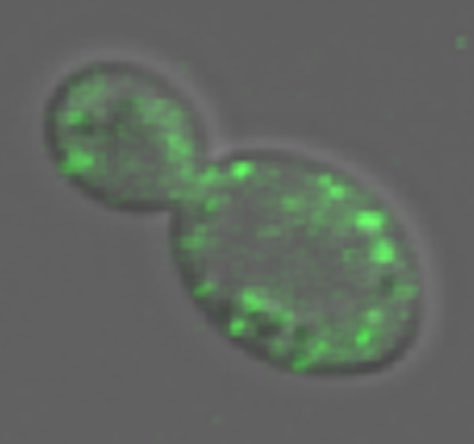
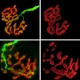
Less is more Green spots in an infected yeast cell are fluorescent prion structures. The size of prion structures is crucial. Smaller structures move more efficiently between cells, so they are more likely to spread disease. Credit: Serio Lab / Brown University
PROVIDENCE, R.I. [Brown University] — Mad Cow disease and its human variant Creutzfeldt—Jakob disease, which are incurable and fatal, have been on a welcome hiatus from the news for years, but because mammals remain as vulnerable as ever to infectious diseases caused by enigmatic proteins called prions, scientists have taken no respite of their own. In the Oct. 29 edition of the journal Science, researchers at Brown University report a key new insight into how prion proteins — the infectious agents — become transmissible: In yeast at least, it is the size of prion complexes, not their number, that determines their efficiency in spreading.
“The dogma in the field was that the misfolding of the protein is sufficient to cause disease, and the clinical course of the infection depended on the amplification of the misfolded protein,” said Tricia Serio, associate professor of molecular biology, cell biology and biochemistry. “But over the years in mammals it has become clear that the abundance of misfolded protein is not a good predictor of disease progression. The question is, What else has to happen for you to get the clinical pathology?”
Cells make prion proteins naturally, although biologists do not understand what their normal role is in mammals. When those proteins misfold in cells, they assemble into structures called aggregates, but other proteins, known as chaperones, attempt to break down the aggregates. The rates at which this assembly and disassembly occurs are determined by the shape or conformation that the prion protein has adopted.
——————————————————————————–
Measuring prion motion
Prions originating in a mother yeast cell (right) are tagged to glow, but are then bleached of their glow in daughter yeast cells (left). That they fade out over time indicates that they are moving back and forth between the cells.
Credit: Serio Lab / Brown University
——————————————————————————–
“Different conformations of the same prion protein can dramatically alter the spread of pathology and the incubation time of prion diseases,” Serio said. “We wanted to learn how.”
By combining experiments in yeast cells with mathematical models, the Brown team found that what affects a prions’ ability to transmit from cell to cell is the size of the structures into which they assemble, Serio said. If the aggregates become too large, they lose their transmissibility among cells. Prion aggregates that remain small are transmitted with greater efficiency.
“In this paper we changed the transmissibility just by shifting the size,” Serio said. “We could change it in either direction.”
The proof was plain to see. Postdoctoral researcher Aaron Derdowski monitored differently sized prion aggregates as they moved among cells under the microscope and could see that smaller ones fared better than larger ones. He also kept track of the spread of different prion structures through a genetic analysis of affected cell populations.
In concert with the experimental work, Suzanne Sindi, a postdoctoral researcher with a joint appointment in molecular biology, cell biology and biochemistry and the Center for Computational Molecular Biology, modeled how cells make and spread prion aggregates, providing a novel simulation of the process that she ran on a computing cluster in the Center for Computation and Visualization at Brown. The model that best replicated experimental observations was the one in which aggregate size, rather than abundance, was the key factor.
Implications for disease
Serio says the insights the team has gained in yeast may better explain what others have observed in mammals as well.
“Previously it was not clear why you would have those outcomes,” she said.
Ultimately the findings could inform future strategies for developing a treatment for prion infection. If researchers unaware of the importance of aggregate size developed a therapy to hinder prion aggregate formation, they might inadvertently make things worse by producing smaller aggregates, Serio said.
“A more effective strategy might be to control the size of the aggregates,” she said, “rather than their presence or absence.”
The findings may also relate to other neurodegenerative diseases that depend on misfolding proteins, such as Alzheimer’s disease or Parkinson’s disease, Serio said.
Other authors on the paper include graduate students Courtney Klaips and Susanne DiSalvo. The National Institutes of Health funded the work.
* The above story is reprinted from materials provided by Brown University
** More information at Brown University (Providence, Rhode Island, USA)