Altered gene tracks RNA editing in neurons.
Altered gene tracks RNA editing in neurons
RNA editing is a key step in gene expression. Scientists at Brown University report in Nature Methods that they have engineered a gene capable of visually displaying the activity of the key enzyme ADAR in living fruit flies.
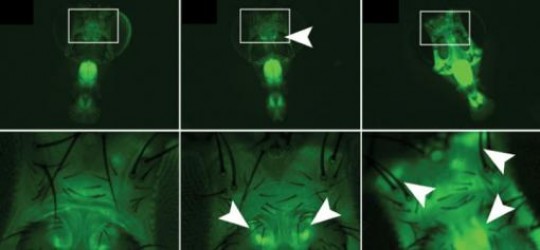 Fluorescing flies/ Fruit flies treated with a gene that glows green in the presence of the RNA-editing enzyme ADAR show significant individual variations (white arrows) where the enzyme is active and how much of it is present. The bottom row enlarges the areas described by white boxes in the top row. /Credit: Reenan Lab/Brown University
Fluorescing flies/ Fruit flies treated with a gene that glows green in the presence of the RNA-editing enzyme ADAR show significant individual variations (white arrows) where the enzyme is active and how much of it is present. The bottom row enlarges the areas described by white boxes in the top row. /Credit: Reenan Lab/Brown University
The advance gives scientists a way to view when and where ADAR is active in a living animal and how much of it is operating. In experiments in fruit flies described in the journal Nature Methods, the researchers show surprising degrees of individual variation in ADAR’s RNA editing activity in the learning and memory centers of the brains of individual flies.
“We designed this molecular reporter to give us a fluorescent readout from living organisms,” said Robert Reenan, professor of biology and senior author of the paper, which appears Dec. 25, 2011. “When it comes to gene expression and regulation, the devil is in the details.”

Robert Reenan/ “We designed this molecular reporter to give us a fluorescent readout from living organisms.”
Biologists already know that errors in transcribing RNA from DNA can lead to improper gene expression in the nervous system and might contribute to diseases such as epilepsy, suicidal depression, and schizophrenia. More recently they’ve gathered evidence that ADAR is associated with disease. For instance in a study in Nature Neuroscience two months ago, Reenan and colleagues at the University of Pennsylvania described profound connections between ADAR and a model of Fragile X mental retardation in fruit flies.
Reenan said that using the new “reporter” tool to look for correlations between ADAR activity levels and behavior or disease might yield new insights into how RNA editing errors lead to such variations. But he also speculated that the mechanics of how he and his research group created the fluorescent ADAR tracking system could be adapted to someday allow therapies based on targeted RNA repair. Their reporter works by requiring ADAR to fix a purposely broken individual letter of RNA on an engineered gene.
“We’re actually repairing RNA at the level of a single informational bit, or nucleotide,” Reenan said. “Here we’ve shown we can take a mutant version of a gene and restore its function, but at the level of RNA rather than DNA.”
A reporter of an editor
Reenan and third author Kyle Jay began working to create the reporter in 2006 when Jay was an undergraduate student just embarking on what would become a celebrated senior thesis at Brown. They started with a well-known tool of molecular biology: a jellyfish gene that produces a protein that glows green upon exposure to ultraviolet light. The strategy was to intentionally break the gene in a way that ADAR is uniquely suited to fix.
First they engineered the gene to include necessary “intron” code that requires a specific splicing operation to take place. Then they inserted the “stop codon” T-A-G in place of T-G-G, which causes transcription to cease, effectively preventing production of the green fluorescent protein. But before splicing occurs and when ADAR finds the stop codon U-A-G in the RNA transcript, it edits the A to an I, which restores the correct information, and translation of the whole gene proceeds as if there were no stop mutation in the DNA. So when splicing and ADAR editing occurs, neurons with the gene reporter glow green.
To see where ADAR editing and splicing were occurring, compared to just splicing alone, they also rigged up an engineered gene with the splicing requirement, but not the T-A-G codon. That would produce yellow fluorescent protein when splicing alone occurred.
Armed with their new ADAR reporter, Reenan and lead author James Jepson set out to make some biological observations in flies. One was that ADAR activity is more pronounced in certain parts of the brains of developing larvae than it is in the brains of adults. The team also found wide variation in ADAR activity in the brains of flies of similar ages from individual to individual. This was a surprise, Reenan said, because all the flies were essentially genetically identical.
A versatile new tool?
Reenan said he is confident that the ADAR reporter could be useful in more organisms than the fruit fly. The idea of creating the reporter grew out of his lab’s studies of comparative genomics in a number of species. ADAR, meanwhile, is found in both invertebrates and vertebrates. In fact, in the paper the researchers describe testing the flexibility of their engineering by inserting into their engineered jellyfish gene — destined as it was for a fruit fly — the splicing intron of a moth.
“Thus it was, a jellyfish-moth gene chimera was crippled by mutation, and repaired by a fruit fly enzyme,” Reenan said. ”Rube Goldberg would be proud.”
Reenan said he plans to use the ADAR reporter in flies to continue the investigation of the genes associated with Fragile X and is eager for someone who works on the disorder in mice to give it a try.
The idea of adapting this method to direct ADAR to fix mistranscribed RNA or reverse DNA damage at the RNA level in a therapeutic fashion is farther into the future. But in a sense, at least ADAR is now on the radar.
In addition to Reenan, Jepson, and Jay, the paper’s other author is Yannis A. Savva. Jepson is also affiliated with Thomas Jefferson University in Philadelphia and Jay now works at the University of California–San Francisco.
An Ellison Medical Foundation Senior Scholar award funded the research.
###
* The above story is adapted from materials provided by Brown University
** More information at Brown University (Providence, Rhode Island, USA)
________________________________________________________________